Monoclonal antibodies are effective drugs against SARS-CoV-2 infection. However, the road is one foot high and the devil is one foot high. After the widespread use of monoclonal antibody drugs, the rapidly mutating new coronavirus has evolved accordingly and has a stronger escape ability.
The latest issue of the international authoritative academic journal "The Lancet-Microbiology" published a communication from the Lyon Hospital in France. The researchers reported that the immunocompromised group received Sotrovimab infusion treatment, the new coronavirus variant strain Omi in the body. Chron has undergone mutation evolution, mutating at sites 340 and 337 of the spike protein, resulting in strong resistance to Sotrovimab.
Sotrovimab, developed by GSK, is a monoclonal antibody for the treatment of patients with severe COVID-19, including those with respiratory, cardiac, metabolic and immunosuppressive comorbidities. A previous study reported that among 100 patients infected with the new coronavirus delta strain (delta, B.1.617.2) and receiving sotrovimab monotherapy, 4 immunocompromised patients had loci 337 or 340 of the spike protein. Drug-resistant mutations developed rapidly.
Sotrovimab neutralizes all beta-coronavirus subgenus sarbecoviruses, including SARS-CoV-2 (SARS-CoV-2), SARS virus, and coronaviruses closely related to bats and pangolins, by binding to highly conserved epitopes within the receptor binding domain. However, the use of specific monoclonal antibodies to target individual viral epitopes requires caution because the virus may rapidly mutate after exposure to these antibodies, resulting in drug resistance. The following mutations are of interest: Mutations at the S:E340K/A/V and S:P337L/T positions were associated with a 100- to 297-fold reduction in sotrovimab neutralization.
Given that sotrovimab is one of the few monoclonal antibodies that remains effective against the widely prevalent Omicron BA.1, monitoring the prevalence of these mutations is critical. From December 2021 to March 2022, as part of routine genome surveillance of the virus at the Lyon Regional Hospital by the French National Centre for Respiratory Viruses, researchers identified 24 of the 18,882 Omicron BA.1 virus genes with the presence of the spike protein. Mutations at sites 340 and 337 occurred at a frequency of 0.13%. One of the 4025 Omicron BA.2 virus genes was found to have mutations at sites 340 and 337 of the spike protein, with a frequency of 0.02%.
The 25 viral gene samples carrying mutations in the spike protein P337 or E340 corresponded to 18 infected individuals. Eight of these patients had clinical data, were immunocompromised and received sotrovimab 0-10 days after symptom onset. For the 6 patients who were followed up, mutations at sites 337 and 340 of the spike protein were absent in their SARS-CoV-2 gene prior to sotrovimab infusion. But 5-18 days after sotrovimab infusion, the new coronavirus in their bodies detected Omicron virus with mutations at sites 337 and 340 of the spike protein at a frequency of 6%-100%.
The researchers say that the emergence of selected resistant viral escape variants is associated with prolonged shedding of SARS-CoV-2, with the longest shedding cycle in these patients reaching 43 days, with only one patient recovering from the infusion on day 24 The infection was completely cleared by the plasma. These results indicate that sotrovimab can rapidly select for mutations at positions 337 and 340 in the BA.1 and BA.2 sublines. These mutated fragments rarely appear in many mutant strains of omicron. According to all 10 042 757 omicron sequences reported in GISAID, the world's largest viral sequence library, only 2756 viral genes have mutations at sites 337 and 340 of spike proteins. , with a frequency of 0.03%. Notably, these mutations were only found in immunocompromised patients treated with sotrovimab. The investigators strongly recommend evaluating monoclonal antibodies as monotherapy in immunocompromised patients because of the risk of escape mutation selection. The researchers say that virological follow-up, including viral sequencing and viral load assessment, should be enhanced in immunocompromised patients receiving monoclonal antibody therapy.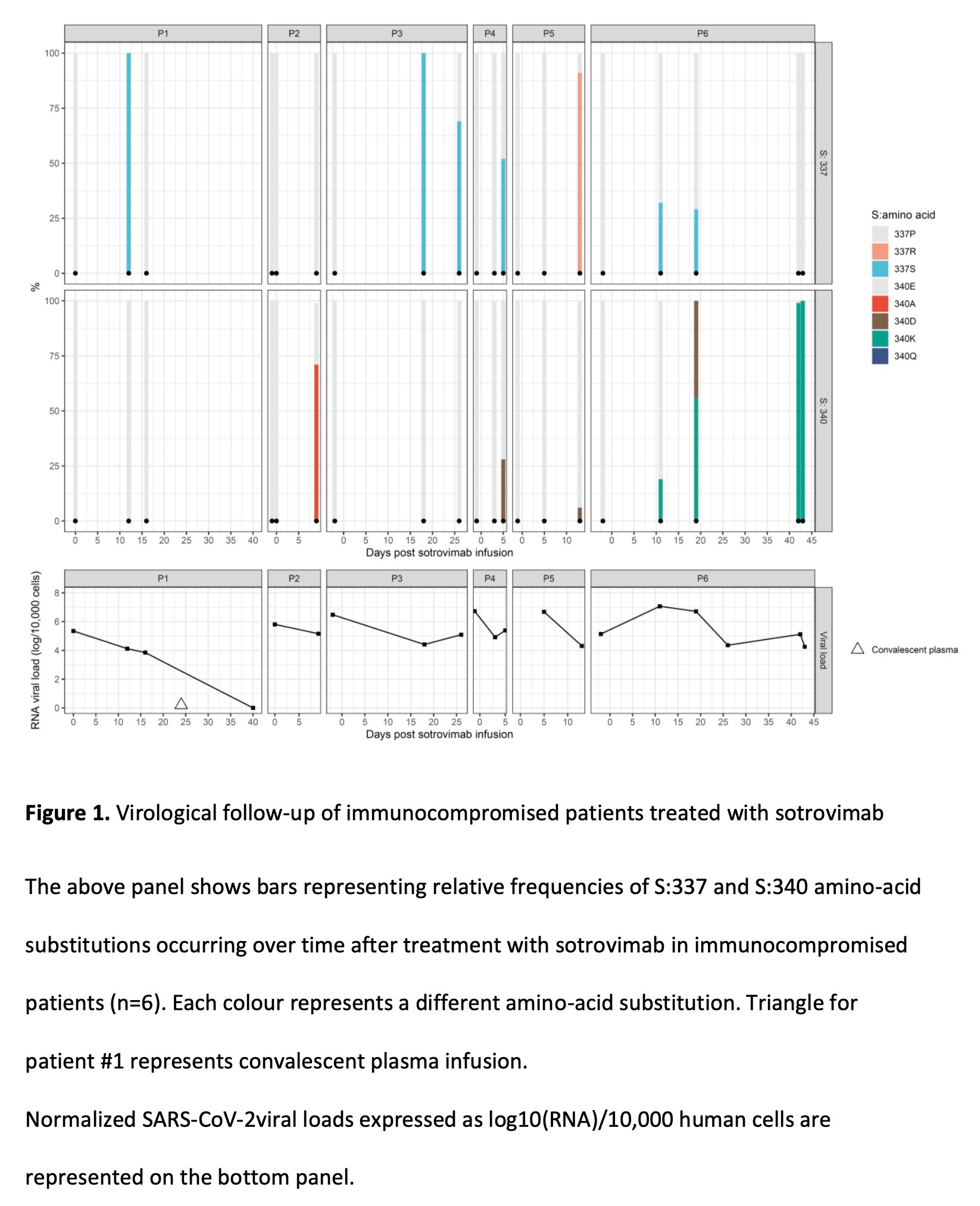
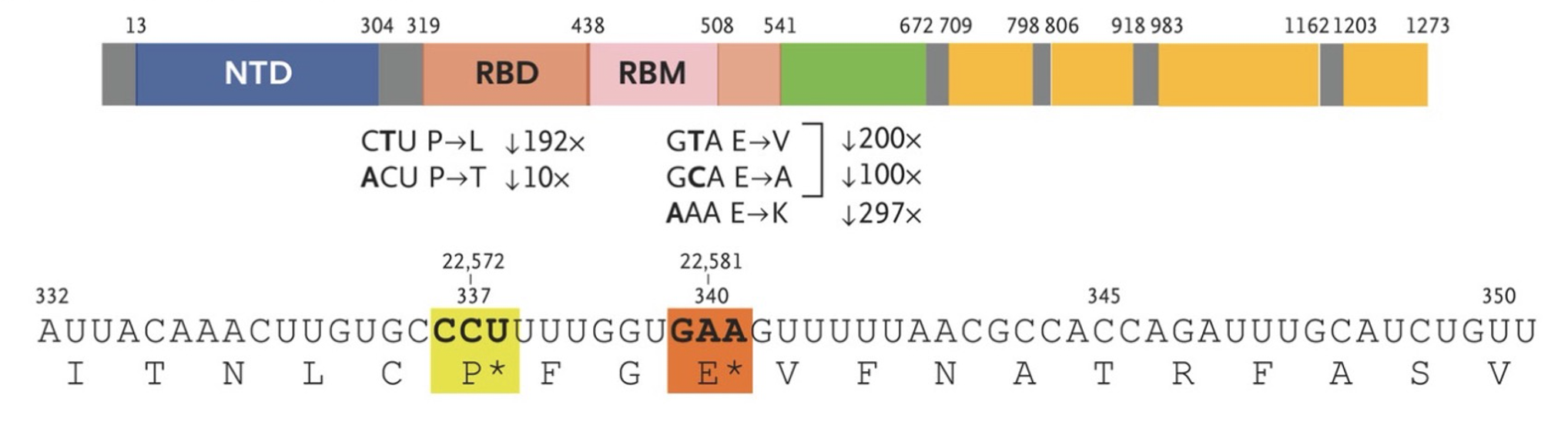 The upper panel shows mutations with high levels of resistance to sotrovimab. Panel A shows the N-terminal domain (NTD), receptor-binding domain (RBD), and receptor-binding motif (RBM) of the SARS-CoV-2 spike protein, and the RBD of the spike protein after sotrovimab treatment. The investigators identified five mutations (S:E340K/A/V and S:P337L/T) that reduced susceptibility to sotrovimab by a factor of 297, 100, 200, 192 and 10, respectively.
The upper panel shows mutations with high levels of resistance to sotrovimab. Panel A shows the N-terminal domain (NTD), receptor-binding domain (RBD), and receptor-binding motif (RBM) of the SARS-CoV-2 spike protein, and the RBD of the spike protein after sotrovimab treatment. The investigators identified five mutations (S:E340K/A/V and S:P337L/T) that reduced susceptibility to sotrovimab by a factor of 297, 100, 200, 192 and 10, respectively.
Previous research published by the Pasteur Institute in France in the journal Nature showed that five antibodies (Bamlanivimab, Etesevimab, Casirivimab, Imdevimab and Regdanvimab) were ineffective against Omi Chron; two antibodies (Cilgavimab and Andintrevimab) were effective against Omi The neutralizing activity of chrones was about 20-fold lower than their activity against delta; only sotrovimab was acceptable and still effective, but the activity against chron omicron was about 3-fold less than that against delta.
The latest issue of the international authoritative academic journal "The Lancet-Microbiology" published a communication from the Lyon Hospital in France. The researchers reported that the immunocompromised group received Sotrovimab infusion treatment, the new coronavirus variant strain Omi in the body. Chron has undergone mutation evolution, mutating at sites 340 and 337 of the spike protein, resulting in strong resistance to Sotrovimab.
Sotrovimab, developed by GSK, is a monoclonal antibody for the treatment of patients with severe COVID-19, including those with respiratory, cardiac, metabolic and immunosuppressive comorbidities. A previous study reported that among 100 patients infected with the new coronavirus delta strain (delta, B.1.617.2) and receiving sotrovimab monotherapy, 4 immunocompromised patients had loci 337 or 340 of the spike protein. Drug-resistant mutations developed rapidly.
Sotrovimab neutralizes all beta-coronavirus subgenus sarbecoviruses, including SARS-CoV-2 (SARS-CoV-2), SARS virus, and coronaviruses closely related to bats and pangolins, by binding to highly conserved epitopes within the receptor binding domain. However, the use of specific monoclonal antibodies to target individual viral epitopes requires caution because the virus may rapidly mutate after exposure to these antibodies, resulting in drug resistance. The following mutations are of interest: Mutations at the S:E340K/A/V and S:P337L/T positions were associated with a 100- to 297-fold reduction in sotrovimab neutralization.
Given that sotrovimab is one of the few monoclonal antibodies that remains effective against the widely prevalent Omicron BA.1, monitoring the prevalence of these mutations is critical. From December 2021 to March 2022, as part of routine genome surveillance of the virus at the Lyon Regional Hospital by the French National Centre for Respiratory Viruses, researchers identified 24 of the 18,882 Omicron BA.1 virus genes with the presence of the spike protein. Mutations at sites 340 and 337 occurred at a frequency of 0.13%. One of the 4025 Omicron BA.2 virus genes was found to have mutations at sites 340 and 337 of the spike protein, with a frequency of 0.02%.
The 25 viral gene samples carrying mutations in the spike protein P337 or E340 corresponded to 18 infected individuals. Eight of these patients had clinical data, were immunocompromised and received sotrovimab 0-10 days after symptom onset. For the 6 patients who were followed up, mutations at sites 337 and 340 of the spike protein were absent in their SARS-CoV-2 gene prior to sotrovimab infusion. But 5-18 days after sotrovimab infusion, the new coronavirus in their bodies detected Omicron virus with mutations at sites 337 and 340 of the spike protein at a frequency of 6%-100%.
The researchers say that the emergence of selected resistant viral escape variants is associated with prolonged shedding of SARS-CoV-2, with the longest shedding cycle in these patients reaching 43 days, with only one patient recovering from the infusion on day 24 The infection was completely cleared by the plasma. These results indicate that sotrovimab can rapidly select for mutations at positions 337 and 340 in the BA.1 and BA.2 sublines. These mutated fragments rarely appear in many mutant strains of omicron. According to all 10 042 757 omicron sequences reported in GISAID, the world's largest viral sequence library, only 2756 viral genes have mutations at sites 337 and 340 of spike proteins. , with a frequency of 0.03%. Notably, these mutations were only found in immunocompromised patients treated with sotrovimab. The investigators strongly recommend evaluating monoclonal antibodies as monotherapy in immunocompromised patients because of the risk of escape mutation selection. The researchers say that virological follow-up, including viral sequencing and viral load assessment, should be enhanced in immunocompromised patients receiving monoclonal antibody therapy.

 The upper panel shows mutations with high levels of resistance to sotrovimab. Panel A shows the N-terminal domain (NTD), receptor-binding domain (RBD), and receptor-binding motif (RBM) of the SARS-CoV-2 spike protein, and the RBD of the spike protein after sotrovimab treatment. The investigators identified five mutations (S:E340K/A/V and S:P337L/T) that reduced susceptibility to sotrovimab by a factor of 297, 100, 200, 192 and 10, respectively.
The upper panel shows mutations with high levels of resistance to sotrovimab. Panel A shows the N-terminal domain (NTD), receptor-binding domain (RBD), and receptor-binding motif (RBM) of the SARS-CoV-2 spike protein, and the RBD of the spike protein after sotrovimab treatment. The investigators identified five mutations (S:E340K/A/V and S:P337L/T) that reduced susceptibility to sotrovimab by a factor of 297, 100, 200, 192 and 10, respectively.Previous research published by the Pasteur Institute in France in the journal Nature showed that five antibodies (Bamlanivimab, Etesevimab, Casirivimab, Imdevimab and Regdanvimab) were ineffective against Omi Chron; two antibodies (Cilgavimab and Andintrevimab) were effective against Omi The neutralizing activity of chrones was about 20-fold lower than their activity against delta; only sotrovimab was acceptable and still effective, but the activity against chron omicron was about 3-fold less than that against delta.
Related Posts
0 Comments
Write A Comments