

 The dead leaf butterfly, scientific name of the dead leaf Nymphalidae, belongs to the genus Nymphalidae, and is a unique type of butterfly in Asia. From the outside, the dead leaf nymph in flight has blue velvet-like wings with bright orange patches on the back of the wings, which looks particularly beautiful. When it stops and closes its wings, its gorgeous wings disappear and become like dead leaves. The pattern of the wings imitates the leaf midrib, secondary vein, petiole and even the decay of the leaf, which is a kind of mimicry (Fig. 1). Mimicry is a means of survival for creatures in nature, which enables creatures to avoid predators as much as possible, and is the result of biological evolution.
The dead leaf butterfly, scientific name of the dead leaf Nymphalidae, belongs to the genus Nymphalidae, and is a unique type of butterfly in Asia. From the outside, the dead leaf nymph in flight has blue velvet-like wings with bright orange patches on the back of the wings, which looks particularly beautiful. When it stops and closes its wings, its gorgeous wings disappear and become like dead leaves. The pattern of the wings imitates the leaf midrib, secondary vein, petiole and even the decay of the leaf, which is a kind of mimicry (Fig. 1). Mimicry is a means of survival for creatures in nature, which enables creatures to avoid predators as much as possible, and is the result of biological evolution.The dead-leaf nymph is the most classic and striking example of leaf mimicry, which the eminent naturalist Alfred Russel Wallace called "the most wonderful and unquestionable protective Mimic". Although more than 150 years have passed since the first observation of these beautiful butterflies, the dead leaf nymph has received considerable attention in taxonomy, morphology, physiology and other disciplines, but research on the genetic basis of their origin and miraculous evolution still lacking.

Figure 1 The picture of the dead leaf nymphalus is provided by the author of the paper
At the macroevolutionary level, based on the whole genome resequencing data of 105 samples of 21 Nymphalidae, combined with methods such as phylogeny, population genetics, population history dynamic modeling, and niche simulation, the study traced the origin and development of Nymphalidae. Evolutionary history. The study found that the early diversification of Nymphalidae coincided with the uplift of the Qinghai-Tibet Plateau in the late Miocene and early Pliocene, when the plateau orogeny formed a significant elevation gradient and diversity in southeastern Tibet and northwestern Yunnan. The climatic conditions in the eastern Himalayas have been very suitable for the survival of Nymphalidae, so the eastern Himalayas may be the habitat of Nymphalidae (Figure 2). differentiation center. The study further found that Nymphalidae species distributed in the Medog area of southeastern Tibet have extensive and directional gene flow to Southeast Asian island species. This result indicates that Nymphalidae species continue to diverge in mainland Asia, and some of these clades Spreading to the eastern and southern island regions, the formation of a land bridge along the southeast coast during the Last Great Glacial Age (LGM) promoted secondary contacts and gene exchanges among species or subspecies of Nymphalidae (Fig. 3). important driver of butterfly diversification.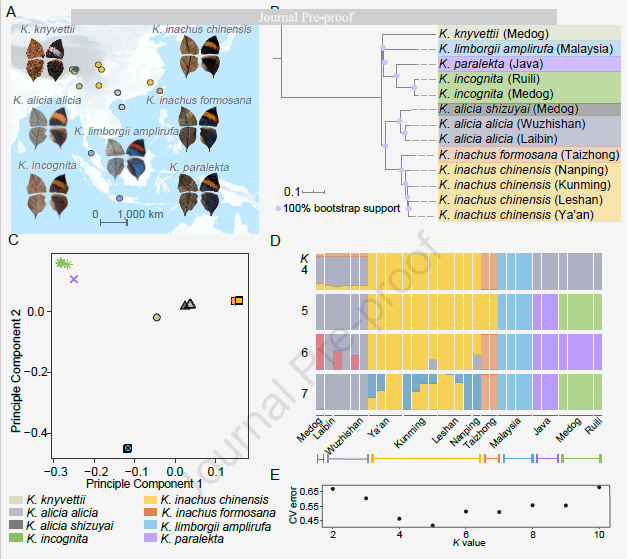
Fig. 2 Geographical distribution, genome-wide phylogenetic relationship and genetic structure of Nymphalus serrata. (A) The dots of different colors represent the sampling sites of different Nymphalidae species, and the corresponding male dorsal and ventral wing patterns are displayed at the same time; (B) The rooted maximum based on the genome-wide SNP data of 14 representative samples Likelihood phylogenetic relationship; (C) Principal component analysis clustering of 34 samples from different species; (D) Population genetic structure of 33 Nymphalidae samples; (E) Cross-validation error plot of population genetic structure shows that, K = 5 is the optimal clustering.
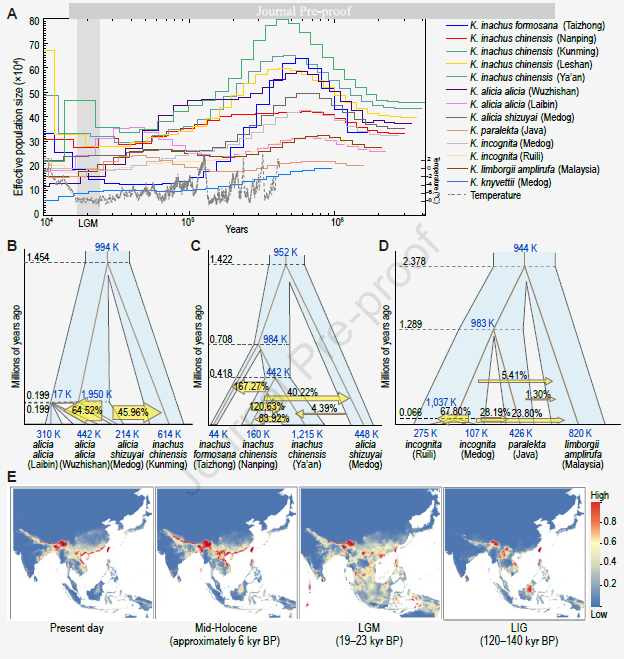
Fig. 3 Population dynamics history of Nymphalidae. (A) Assuming mutation rate μ = 3 × 10-9 and average generation length g = 0.5 years, the historical effective population size inferred by PSMC; (B, C, D) The differentiation time of Nymphalidae inferred by G-PhoCS , effective population size and migration rate, the dotted line indicates the differentiation time, and the clade width indicates the proportion of population size; (E) Using Maxent to simulate the niche of Nymphalidae, predicting suitable habitats from the last interglacial period (LIG), color matching From red to blue, the environmental suitability is from high to low.
At the microevolutional level, the study constructed a hybrid line of different wing types of Nymphaea sinensis, and found 10 leaf mimic phenotypes . Genome-wide association analysis identified a wing phenotype regulatory locus, the cortex gene. Comprehensive evidence obtained through high-quality genome sequencing, chromatin interaction, and gene editing suggests that this gene plays an important role in regulating leaf-mimicking wing-type polymorphisms, and it may be involved in wing pigmentation and wing pigmentation by regulating potential downstream genes snake. Color conversion. Transcriptional-level findings also revealed the spatiotemporal regulation of cortex gene expression in developmental stages and tissue morphology (Figure 4). Further, population genetic analysis found that leaf mimicry was maintained by balanced selection, which may be a negative frequency-dependent selection with clear adaptive value, supporting the role of long-term balanced selection in adaptive evolution. The results provide clear evidence that cortex gene plays a key role in regulating leaf mimicry formed by natural selection, and its structural and non-structural genetic mechanisms are conducive to maintaining the diversity of leaf mimicry.Diversity evolution of Nymphalus species is a complex and attractive model, and its in-depth study reveals that complex evolutionary innovations are driven by both environmental geography and natural selection , and provide insights for the study of natural selection to maintain diversity. a unique perspective.
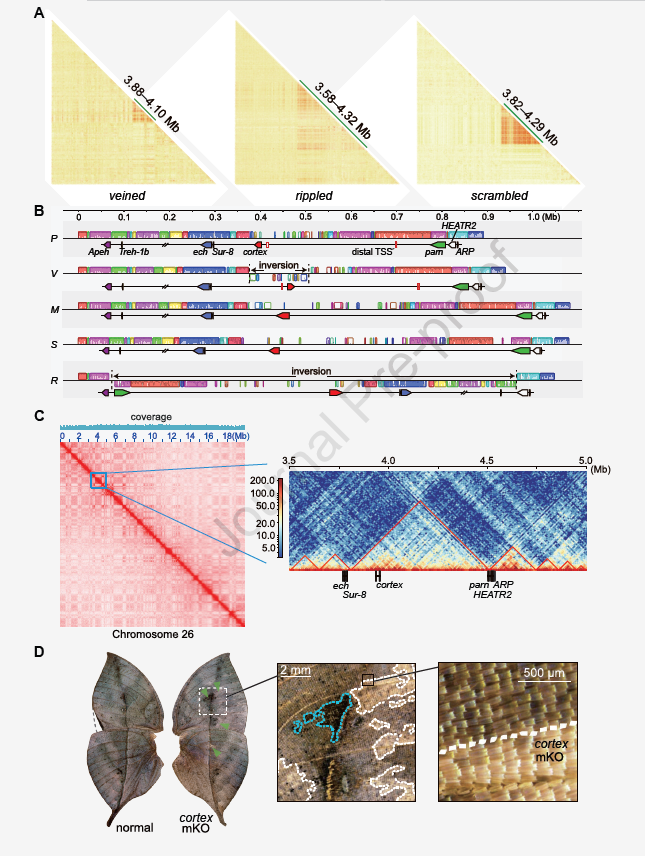
Figure 4 Suppression of cortex haplotype recombination and functional verification of cortex gene in Nymphaea sinensis. (A) Linkage disequilibrium plotted with r2, and elevated linkage disequilibrium observed in cortex regions of veined, corrugated, and scrambled mimic samples; (B) five haplotype cortex regions aligned to reveal V and R Haplotypes for two different chromosomal inversions; (C) Heatmap showing the average interaction frequency of all putative topological binding domains (TADs) on chromosome 26, and putative TADs containing cortex and adjacent genes in the S haplotype (D) The ventral wing surface shows cortex loss-of-function effects triggered by CRISPR/Cas9 genome editing, cortex knockout results in faded scale pigmentation around the lateral veins and blurring of the ventral midrib.
AbstractThe dead leaf butterfly belongs to the Nymphalidae family, and its wings have phenotypic polymorphisms, which can disguise the shape of dead leaves. This iconic protective mimicry provides an interesting evolutionary paradigm for studying biodiversity. This study integrates multi-omics data analysis and functional verification methods to infer the evolutionary history of Nymphalidae species and explore the genetic basis for the diverse leaf mimicry of these species' wings. The study found that Nymphalus species diversified in the eastern Himalayas and spread to East and Southeast Asia. The polymorphism of wing-leaf mimicry is regulated by the wing-type gene cortex, which is fixed in Nymphaea species by long-term balanced selection. This study provides insights into the macroevolutionary and microevolutionary processes of model species derived from mountain ecosystems.
Reviewer Profile

Jiang Dechun, Young Researcher of Chengdu Institute of Biology, Chinese Academy of Sciences, Member of Youth Innovation Promotion Association of Chinese Academy of Sciences
Jiang Dechun, Ph.D., is a young researcher at the Chengdu Institute of Biology, Chinese Academy of Sciences, and a member of the Youth Innovation Promotion Association of the Chinese Academy of Sciences in 2021. Mainly engaged in the research on the evolution of amphibian diversity, exploring the molecular phylogenetic relationship of amphibians, evolution of species characteristics, biogeography, etc., focusing on the evolution of amphibian geographical pattern and the evolution mechanism of environmental adaptation. At present, he has published more than 20 SCI papers in National Science Review, PNAS and other journals.about the author

Zhang Wei, Ph.D., is a researcher and doctoral supervisor of the School of Life Sciences of Peking University and the Peking University-Tsinghua Joint Center for Life Sciences.
Researcher Zhang Wei, received a Bachelor of Science degree from Shandong University in 2005 and a Ph.D. degree from Peking University in 2011. From 2012 to 2017, he did post-doctoral research at the University of Chicago, USA. From 2018 to present, he has been working in the School of Life Sciences of Peking University and the Peking University-Tsinghua Joint Center for Life Sciences. In the same year, he joined the State Key Laboratory of Protein and Plant Gene Research and the Ecological Research Center of Peking University. By integrating research methods such as population genetics, evolutionary development, gene editing, and biological information, researcher Zhang Wei has comprehensively and systematically studied the evolution of butterfly mimicry, a classic macro-evolutionary example, including leaf mimicry, Bayesian mimicry, Mushi mimicry, and warning color. genetic mechanism. Published results in professional journals such as Cell, Nature, Nature Communications, Science Advances, Genome Biology, etc. as the corresponding author or the first author (including co-authors); research results have been included in two textbooks; recommended by Faculty 1000; written by Nature magazine Invited commentary; has been reported many times by Nature magazine and Science magazine. In 2019, he was awarded the title of "Boya Young Scholar" of Peking University and an excellent instructor for undergraduate graduation design in ordinary colleges and universities in Beijing. He presided over the General Program of National Natural Science Foundation of China and the Outstanding Youth Program of Beijing Natural Science Foundation.The current research interest of the research group is to integrate experimental and computational biology research methods to explore the theoretical basis and molecular mechanisms of important evolutionary problems, such as speciation, adaptive evolution, and infiltration evolution. Our research subjects are often characterized by species abundance, adaptive radiation, and incomplete reproductive isolation. For example, lepidopteran insects are rich in species, have extensive interspecific hybridization, and have diverse wing patterns, which are classic systems for studying adaptive evolution. The rapid development of sequencing technology and gene editing technology has made it possible to perform evolutionary genomics and population genetics studies at the genome-wide level and to perform functional identification of target genes for non-model organisms. The main research directions include: the origin and genetic mechanism of leaf mimicry; the male and female dimorphism during the development of Lepidoptera; the identification of adaptive gene introgression, functional research and evolutionary models, etc.
(Original title "Cell Cover Study: Mimetic Diversity and Evolution of Nymphalidae | Author Interview and Review of Cell Press Youth Promotion Association")
