
Omicron subtype BA.2 is currently the main subtype of SARS-CoV-2 infection worldwide. However, some recent data suggest that its offspring BA.2.12.1 and BA.4 and BA.5 have soared to dominate the US and South Africa respectively.
Will these new variants replace BA.2, and how will they disrupt current prevention and treatment strategies once they become the global mainstream strain? A team led by David D. Ho, director of the Aaron Diamond AIDS Research Center in the College of Physicians and Surgeons at Columbia University's Vagelos College of Medicine and a foreign academician at the Chinese Academy of Engineering, investigated these surges. Subvariants were subjected to a systematic antigenic analysis.
They concluded that BA.2.12.1 was 1.8-fold more resistant to vaccinated and boosted sera than BA.2, while BA.4/5 was much more resistant to 4.2-fold and thus more likely to lead to breakthrough Sexual infection, that is, getting infected despite having an existing vaccine . Furthermore, of the currently approved therapeutic antibodies for clinical use, only bebtelovimab (LY-COV1404) remains fully potent against BA.2.12.1 and BA.4/5.
The study argues that the Omicron lineage of SARS-CoV-2 continues to evolve, and these resulting sub-variants are not only more infectious but also more prone to antibody escape.
He Dayi is the founder of the Allen Diamond AIDS Research Center, a foreign academician of the Chinese Academy of Engineering, a member of the American Academy of Medicine, the American Academy of Sciences, and the American Academy of Arts and Sciences. He invented the "cocktail therapy" in the 1990s to treat AIDS. During the COVID-19 pandemic, his team has been tracking the mutation of the new coronavirus for a long time and assessing the impact on vaccines and antibodies in a timely manner, and has previously published several important studies. The latest research paper, published on the preprint site bioRxiv, has not yet been peer-reviewed.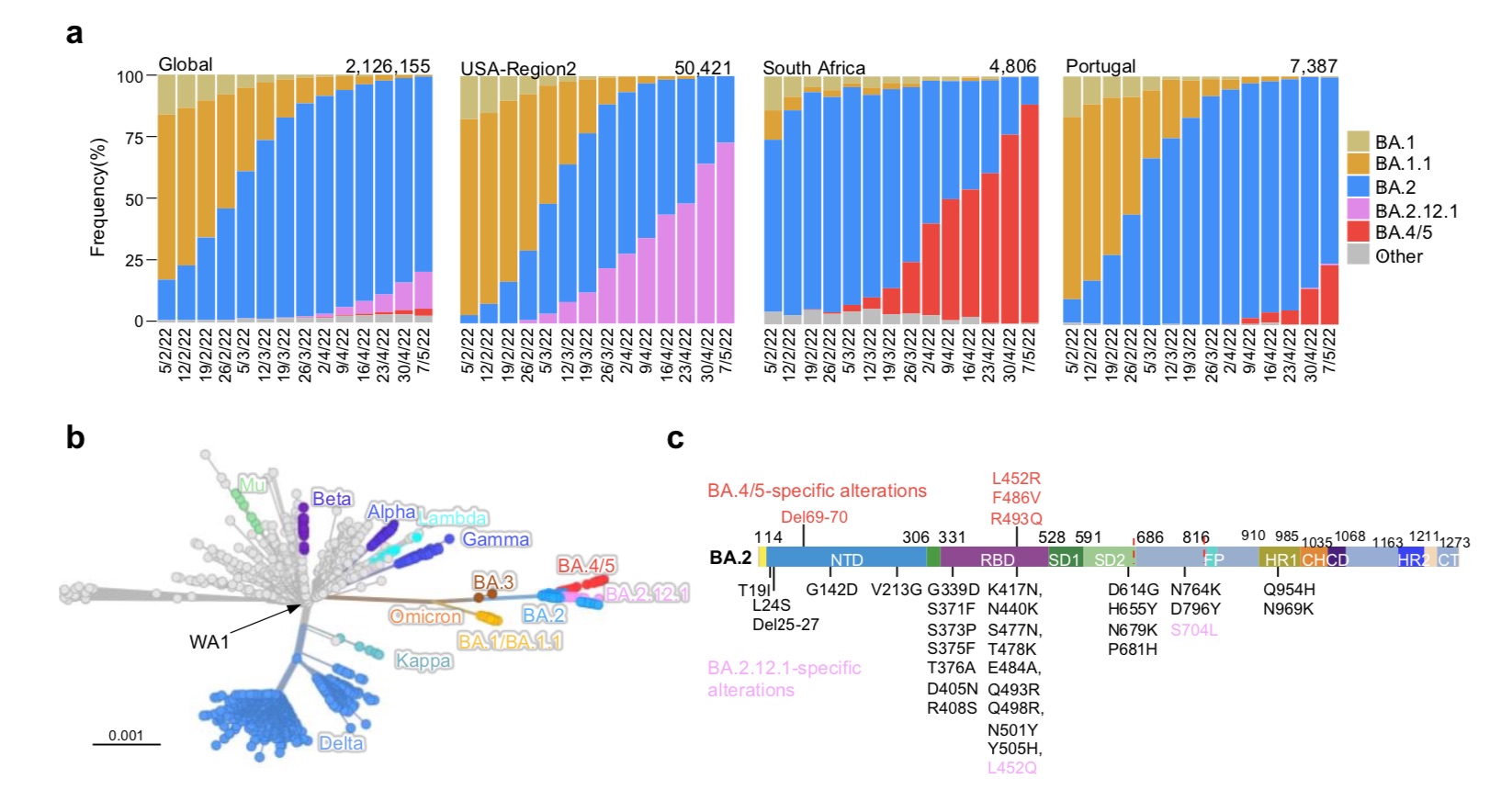
He et al. mentioned that these new omicron sub-variants have been detected worldwide, although the overall prevalence is currently low. However, their growth trajectories in the United States and South Africa suggest that these strains have significant transmission advantages that could lead to further expansion.
Phylogenetically, these new subvariants evolved independently of BA.2. The spike protein of BA.2.12.1 contains L452Q and S704L mutations in addition to the known BA.2 mutations. While the spike proteins of BA.4 and BA.5 are identical, both have 4 additional mutations, namely Del69-70, L452R, F486V and R493Q. Among them, R493Q is a reversion mutation (reversion mutation, re-mutation and restoring the original gene).
The location of these mutations in the RBD (receptor domain) of the spike protein raises concerns that BA.2.12.1 and BA.4/5 may have evolved to further escape neutralizing antibodies. The RBD is the region where the viral spike protein binds to the receptor on infected cells, ACE2 (angiotensin-converting enzyme 2), and is therefore of particular interest.
To understand the antigenic differences between BA.2.12.1 and BA.4/5 from the previous Omicron subvariants (BA.1, BA.1.1 and BA.2) and wild-type SARS-CoV-2 (D614G), The research team prepared pseudoviruses and then assessed the susceptibility of each pseudovirus to neutralization with 21 monoclonal antibodies (mAbs). There are 19 proteins targeting 4 epitopes of RBD, including REGN10987 (imdevimab), REGN10933 (casirivimab), COV2-2196 (tixagevimab), COV2-2130 (cilgavimab), LY-CoV555 (bamlanivimab), CB6 (etesevimab) , Brii-196 (amubarvimab), Brii-198 (romlusevimab), S309 (sotrovimab), LY-CoV1404 (bebtelovimab), ADG-2, DH1047, S2X259, CAB-A17 and ZCB11, as well as 1- 20, 2-15, 2-7 and 10-40. Two other monoclonal antibodies, 4-18 and 5-7, target the N-terminal domain (NTD).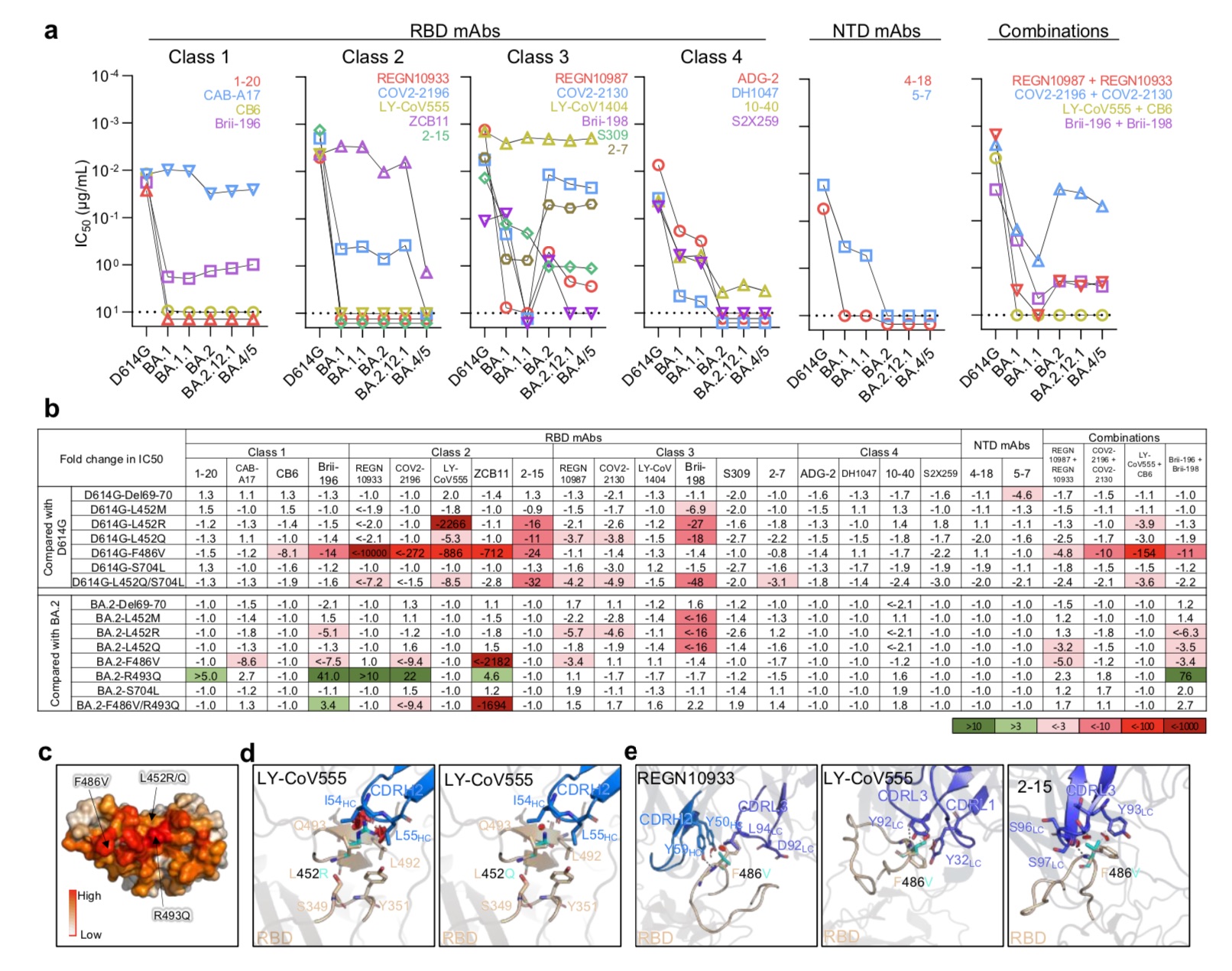
The study found that only four mAbs, CAB-A17, COV2-2130, 2-7 and LY-COV1404, still maintained good in vitro titers against BA.2.12.1 and BA.4/5.
But importantly, among these 4 mAbs, only LY-COV1404 (bebtelovimab) has been authorized for clinical treatment. Bebtelovimab was granted Emergency Use Authorization (EUA) by the U.S. FDA on February 11, 2022 for the treatment of adults and pediatric patients (at least 40 kg) with mild to moderate COVID-19, as well as those with high People at risk of developing severe COVID-19.
In addition, the in vitro neutralizing activity against both BA.2.12.1 and BA.4/5 was significantly reduced for antibody combinations previously authorized or approved for clinical use.
The research team also concluded that M, R, and Q substitutions of the L452 residue previously found in Delta and Lambda variants largely confer resistance to class 2 and 3 RBD mAbs, with L452R being the more deleterious one. mutation. In addition, F486V also broadly attenuated the neutralizing activity of several class 1 and 2 RBD mAbs. Notably, the mutation reduced ZCB11's potency by more than 2,000-fold.
In contrast, the back mutation R493Q sensitized BA.2 to several class 1 and class 2 RBD monoclonal antibodies and could be neutralized. This finding is also in line with previous work by the research team showing that Q493R, found in an early Omicron subvariant, mediated resistance to the same set of mAbs in this study.
The research team also conducted further studies on the affinity of these proliferating new variants. They point out that epidemiological data clearly show that both BA.2.12.1 and BA.4/5 are highly infectious; however, as some teams have reported, some additional mutations in these subvariant RBDs also increase the virus Possibility of significant loss of affinity for the receptor, human ACE2 (hACE2).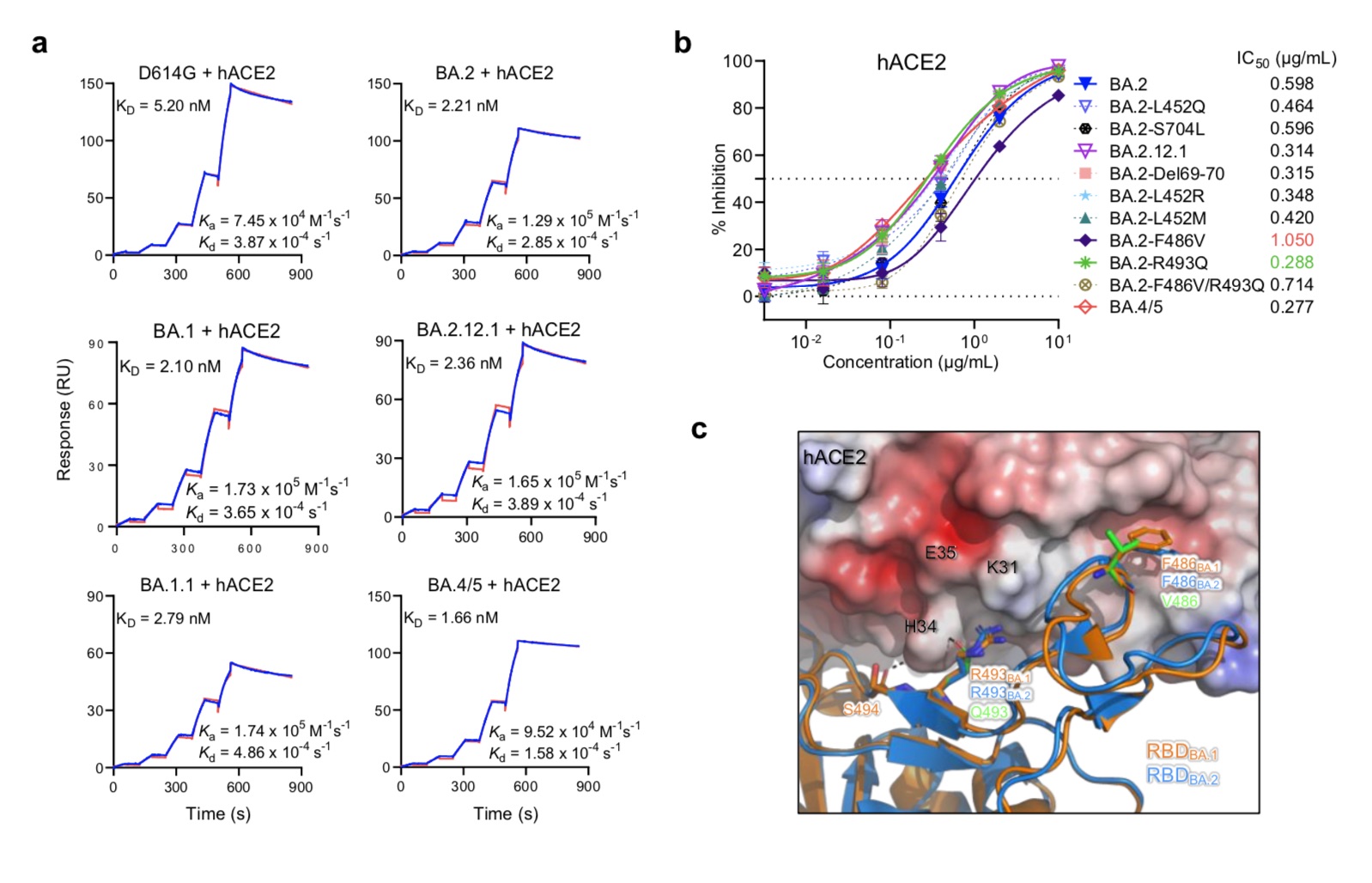
Their results also showed that the F486V mutation decreased receptor affinity, but the R493Q back mutation increased receptor affinity. Interestingly, before the emergence of BA.4/5, the mutation frequency of F486 was very low (less than 10E-5), which may be due to the reduced receptor affinity.
Next, the research team also assessed the degree of neutralization resistance of BA.2.12.1 and BA.4/5 to sera from 4 different clinical cohorts. Given that two-dose mRNA vaccinators were mostly unable to neutralize the early Omicron subvariants, their 4 cohorts involved three-dose mRNA vaccine recipients (booster), non-Omicron pre-infection, or infection. Post-vaccination, post-vaccination BA.1 breakthrough infection, post-vaccination BA.2 breakthrough infection.
For the booster needle group, the neutralizing titers of BA.1, BA.1.1, and BA.2 were significantly lower (4.6-fold to 6.2-fold) compared with D614G (Fig. 4b). BA.2.12.1 (8.1 times) and BA.4/5 (19.2 times) are even lower. Similar serum neutralization trends were observed in other cohorts.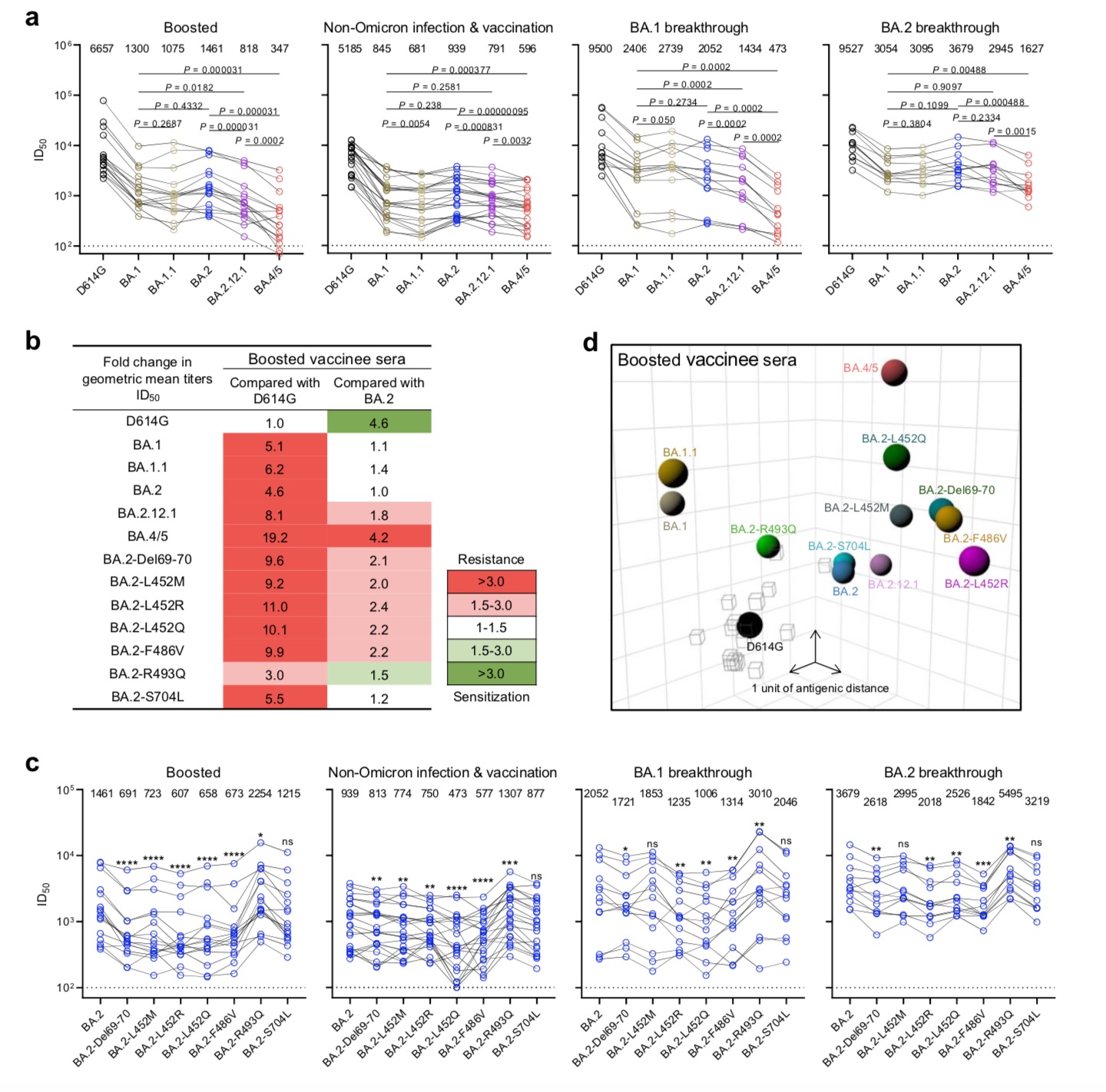
Overall, the team believes that this map clearly shows that BA.4/5 is more resistant to neutralization by sera obtained from vaccinated and boosted individuals, some of which are mutations that lead to antibody evasion.
He Dayi and others pointed out that a key question now is whether BA.4 and BA.5 will defeat BA.2.12.1? They believe that the answer will soon be revealed in the "battlefield".
The research team believes that from an epidemiological point of view, because these two omicron sub-variants have a clear advantage in transmission, despite the many mutations in the spike protein, their interaction with the hACE2 receptor is not obvious. It's no surprise that binding remains strong. In fact, the affinity of BA.4/5 for the receptor may be slightly higher.
As the Omicron lineage has evolved over the past few months, each successive subvariant appears to have spread faster and faster in humans and has become stronger at evading antibodies, the team concluded. They stressed that we must note that each major global variant of SARS-CoV-2 appeared randomly and unexpectedly, "We remain vigilant and strive to do a good job of collective surveillance."
Will these new variants replace BA.2, and how will they disrupt current prevention and treatment strategies once they become the global mainstream strain? A team led by David D. Ho, director of the Aaron Diamond AIDS Research Center in the College of Physicians and Surgeons at Columbia University's Vagelos College of Medicine and a foreign academician at the Chinese Academy of Engineering, investigated these surges. Subvariants were subjected to a systematic antigenic analysis.
They concluded that BA.2.12.1 was 1.8-fold more resistant to vaccinated and boosted sera than BA.2, while BA.4/5 was much more resistant to 4.2-fold and thus more likely to lead to breakthrough Sexual infection, that is, getting infected despite having an existing vaccine . Furthermore, of the currently approved therapeutic antibodies for clinical use, only bebtelovimab (LY-COV1404) remains fully potent against BA.2.12.1 and BA.4/5.
The study argues that the Omicron lineage of SARS-CoV-2 continues to evolve, and these resulting sub-variants are not only more infectious but also more prone to antibody escape.
He Dayi is the founder of the Allen Diamond AIDS Research Center, a foreign academician of the Chinese Academy of Engineering, a member of the American Academy of Medicine, the American Academy of Sciences, and the American Academy of Arts and Sciences. He invented the "cocktail therapy" in the 1990s to treat AIDS. During the COVID-19 pandemic, his team has been tracking the mutation of the new coronavirus for a long time and assessing the impact on vaccines and antibodies in a timely manner, and has previously published several important studies. The latest research paper, published on the preprint site bioRxiv, has not yet been peer-reviewed.

Prevalence of the SARS-CoV-2 Omicron subvariant.
BA.2.12.1 emerged in the United States in early February and expanded significantly in the northeastern United States, including New York, where BA.2.12.1 now accounts for 70% of all new SARS-CoV-2 infections in the region above. BA.4 and BA.5 emerged in South Africa in January and quickly became the dominant strains, with a combined share of over 88%.He et al. mentioned that these new omicron sub-variants have been detected worldwide, although the overall prevalence is currently low. However, their growth trajectories in the United States and South Africa suggest that these strains have significant transmission advantages that could lead to further expansion.
Phylogenetically, these new subvariants evolved independently of BA.2. The spike protein of BA.2.12.1 contains L452Q and S704L mutations in addition to the known BA.2 mutations. While the spike proteins of BA.4 and BA.5 are identical, both have 4 additional mutations, namely Del69-70, L452R, F486V and R493Q. Among them, R493Q is a reversion mutation (reversion mutation, re-mutation and restoring the original gene).
The location of these mutations in the RBD (receptor domain) of the spike protein raises concerns that BA.2.12.1 and BA.4/5 may have evolved to further escape neutralizing antibodies. The RBD is the region where the viral spike protein binds to the receptor on infected cells, ACE2 (angiotensin-converting enzyme 2), and is therefore of particular interest.
To understand the antigenic differences between BA.2.12.1 and BA.4/5 from the previous Omicron subvariants (BA.1, BA.1.1 and BA.2) and wild-type SARS-CoV-2 (D614G), The research team prepared pseudoviruses and then assessed the susceptibility of each pseudovirus to neutralization with 21 monoclonal antibodies (mAbs). There are 19 proteins targeting 4 epitopes of RBD, including REGN10987 (imdevimab), REGN10933 (casirivimab), COV2-2196 (tixagevimab), COV2-2130 (cilgavimab), LY-CoV555 (bamlanivimab), CB6 (etesevimab) , Brii-196 (amubarvimab), Brii-198 (romlusevimab), S309 (sotrovimab), LY-CoV1404 (bebtelovimab), ADG-2, DH1047, S2X259, CAB-A17 and ZCB11, as well as 1- 20, 2-15, 2-7 and 10-40. Two other monoclonal antibodies, 4-18 and 5-7, target the N-terminal domain (NTD).

Resistance of Omicron subvariants to neutralization by monoclonal antibodies.
Overall, 18 mAbs completely or partially lost their neutralizing activity against BA.2.12.1, and 19 mAbs completely or partially lost their neutralizing activity against BA.4/5. From the comparison of BA.2 and BA.2.12.1, except for 3 class 3 (Class 3) RBD monoclonal antibodies (Brii-198, REGN10987 and COV2-2130) that are inactive or have further effects on BA.2.12.1, The neutralization spectrum of the two is similar. Compared with BA.2 and BA.2.12.1, BA.4/5 showed significant neutralizing resistance to two class 2 RBD monoclonal antibodies (ZCB11 and COV2-2196), and was resistant to two class 3 RBD monoclonal antibodies. Antibodies (REGN10987 and COV2-2130) showed moderate resistance.The study found that only four mAbs, CAB-A17, COV2-2130, 2-7 and LY-COV1404, still maintained good in vitro titers against BA.2.12.1 and BA.4/5.
But importantly, among these 4 mAbs, only LY-COV1404 (bebtelovimab) has been authorized for clinical treatment. Bebtelovimab was granted Emergency Use Authorization (EUA) by the U.S. FDA on February 11, 2022 for the treatment of adults and pediatric patients (at least 40 kg) with mild to moderate COVID-19, as well as those with high People at risk of developing severe COVID-19.
In addition, the in vitro neutralizing activity against both BA.2.12.1 and BA.4/5 was significantly reduced for antibody combinations previously authorized or approved for clinical use.
The research team also concluded that M, R, and Q substitutions of the L452 residue previously found in Delta and Lambda variants largely confer resistance to class 2 and 3 RBD mAbs, with L452R being the more deleterious one. mutation. In addition, F486V also broadly attenuated the neutralizing activity of several class 1 and 2 RBD mAbs. Notably, the mutation reduced ZCB11's potency by more than 2,000-fold.
In contrast, the back mutation R493Q sensitized BA.2 to several class 1 and class 2 RBD monoclonal antibodies and could be neutralized. This finding is also in line with previous work by the research team showing that Q493R, found in an early Omicron subvariant, mediated resistance to the same set of mAbs in this study.
The research team also conducted further studies on the affinity of these proliferating new variants. They point out that epidemiological data clearly show that both BA.2.12.1 and BA.4/5 are highly infectious; however, as some teams have reported, some additional mutations in these subvariant RBDs also increase the virus Possibility of significant loss of affinity for the receptor, human ACE2 (hACE2).

Affinity of the omicron sub-variant spike protein for human ACE2.
In this latest systematic study, the research team found that the KD value of BA.4/5 was 1.66 nM by measuring the binding affinity of the purified D614G spike protein and the main Omicron sub-variant to dimeric hACE2, The KD value of BA.2.12.1 was 2.36nM, and the KD value of BA.1.1 was 2.79nM. Impressively, although more than 17 mutations in the RBDs of BA.2.12.1 and BA.4/5 helped them evade antibody neutralization, these variants also evolved at the same time, resulting in more KD 5.20 nM) higher receptor affinity.Their results also showed that the F486V mutation decreased receptor affinity, but the R493Q back mutation increased receptor affinity. Interestingly, before the emergence of BA.4/5, the mutation frequency of F486 was very low (less than 10E-5), which may be due to the reduced receptor affinity.
Next, the research team also assessed the degree of neutralization resistance of BA.2.12.1 and BA.4/5 to sera from 4 different clinical cohorts. Given that two-dose mRNA vaccinators were mostly unable to neutralize the early Omicron subvariants, their 4 cohorts involved three-dose mRNA vaccine recipients (booster), non-Omicron pre-infection, or infection. Post-vaccination, post-vaccination BA.1 breakthrough infection, post-vaccination BA.2 breakthrough infection.
For the booster needle group, the neutralizing titers of BA.1, BA.1.1, and BA.2 were significantly lower (4.6-fold to 6.2-fold) compared with D614G (Fig. 4b). BA.2.12.1 (8.1 times) and BA.4/5 (19.2 times) are even lower. Similar serum neutralization trends were observed in other cohorts.

BA.2.12.1 and BA.4/5 exhibited larger serum neutralization resistance profiles relative to D614G and BA.2.
The research team also constructed a map of the antigenic distances between D614G, various Omicron sub-variants, and individual point mutations using samples from the booster group. The antigen map shows that BA.1, BA.1.1 and BA.2 are approximately equidistant from D614G, approximately 3-4 antigenic units each. BA.2.12.1 is 1 antigenic unit apart from BA.2. Most strikingly, BA.4/5 is 6 antigenic units away from D614G and 4 antigenic units away from BA.2. In addition, the point mutants Del69-70, L452M/Q/R and F486V all increased the antigenic distance to BA.2 and D614G, whereas R493Q did the opposite.Overall, the team believes that this map clearly shows that BA.4/5 is more resistant to neutralization by sera obtained from vaccinated and boosted individuals, some of which are mutations that lead to antibody evasion.
He Dayi and others pointed out that a key question now is whether BA.4 and BA.5 will defeat BA.2.12.1? They believe that the answer will soon be revealed in the "battlefield".
The research team believes that from an epidemiological point of view, because these two omicron sub-variants have a clear advantage in transmission, despite the many mutations in the spike protein, their interaction with the hACE2 receptor is not obvious. It's no surprise that binding remains strong. In fact, the affinity of BA.4/5 for the receptor may be slightly higher.
As the Omicron lineage has evolved over the past few months, each successive subvariant appears to have spread faster and faster in humans and has become stronger at evading antibodies, the team concluded. They stressed that we must note that each major global variant of SARS-CoV-2 appeared randomly and unexpectedly, "We remain vigilant and strive to do a good job of collective surveillance."
Related Posts
0 Comments
Write A Comments